Research Highlights
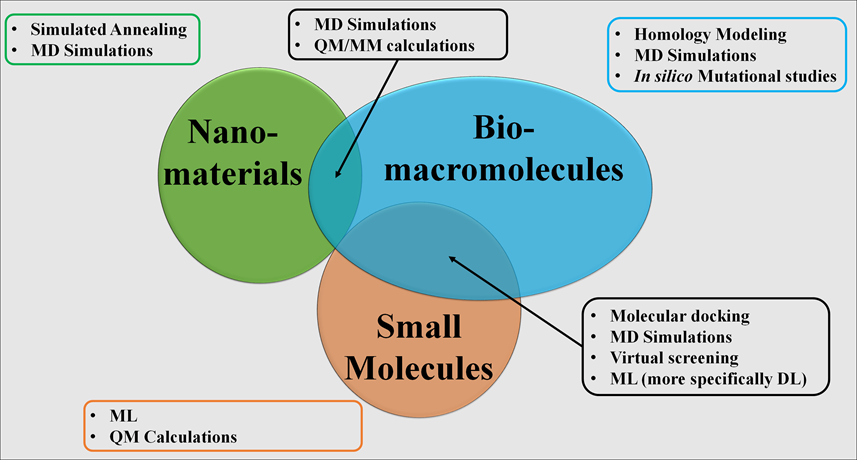
My research background in pharmacoinformatics, integrating the pharmaceutical sciences with computational biology and computational chemistry, has empowered me to make pioneering contributions to the research field. This discipline inspires me by enabling an atomic-level mechanistic understanding of therapeutic actions and toxicity. The integration of molecular modeling and machine learning (ML) is revolutionizing the design, discovery, and development paradigm, enabling predictive insights and optimizing outcomes. Participating in this cutting-edge discipline motivates me to advance scientific knowledge and develop innovations with profound implications across multiple domains, particularly within healthcare and nanoscience. My work has focused on the interplay of (a) nanomaterials and biomacromolecules, particularly in investigating the interactions between pharmacologically relevant protein targets and widely occurring nanomaterials, and (b) biomacromolecules and small molecules to investigate the mechanism of pharmacological actions and identify novel therapeutic agents.
My research focuses on the dynamic intersection of computational science and molecular biology, where I have worked to unravel the complex dynamics of nanoparticle-biomolecule interactions and the intricate mechanisms underlying biomolecular interactions with small molecules. I have successfully applied my computational biology and computational chemistry expertise to uncover several groundbreaking insights. My interdisciplinary expertise in pharmaceutical sciences and informatics provides a solid foundation for tackling challenging questions across diverse scientific domains. With years of experience in state-of-the-art computational methodologies, such as machine learning and molecular dynamics simulations, I am driven to uncover transformative insights and develop predictive models that advance our understanding of biomolecular systems. This unique combination of computational excellence and domain-specific innovation positions me to push the boundaries of computational research in nano and biomolecular sciences, paving the way for groundbreaking discoveries.
Nanomaterials and Environmental Impact
In this domain, my research focuses on leveraging computational modeling to uncover the intricate behaviors of nanomaterials, including their molecular assembly, surface properties, crystallization dynamics, and interactions with biologically significant molecules. By evaluating the binding potential of nanoparticles with various amino acids and protein surfaces, a deeper dive into their biological implications is possible. In light of recent findings highlighting the blood-brain barrier (BBB) breaching potential of plastics, studying the affinity of nanoplastics with neuropharmacologically relevant proteins becomes important. Our recent publication (https://doi.org/10.1021/acs.biomac.4c00918) focuses on the intermolecular communication between polyethylene-based nanoplastics and human α-synuclein (hαSn), a pathophysiologically significant protein and key biomarker in various neurodegenerative disorders. We have shown, for the first time a bidirectional impact of nanoplastics on proteins, through molecular dynamics simulations.
Click on the videos to play
Rapid adsorption of human α synuclein on polyethylene plastic surface
Adsorption of multiple human α synuclein on polyethylene plastic surface
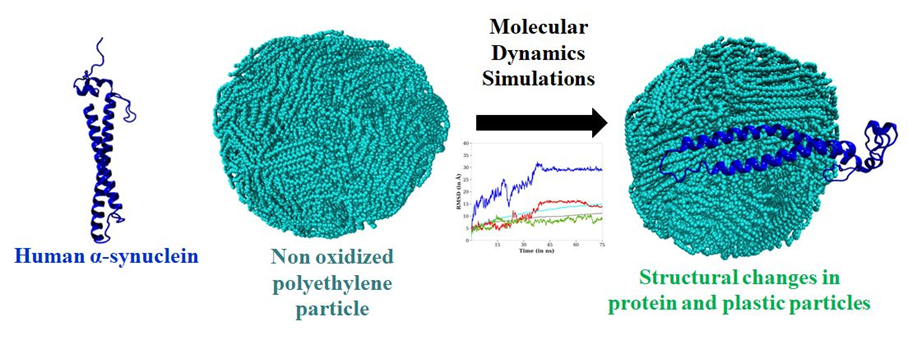
The human α synuclein adsorption on polyethylene nanoplastic surface (https://doi.org/10.1021/acs.biomac.4c00918)
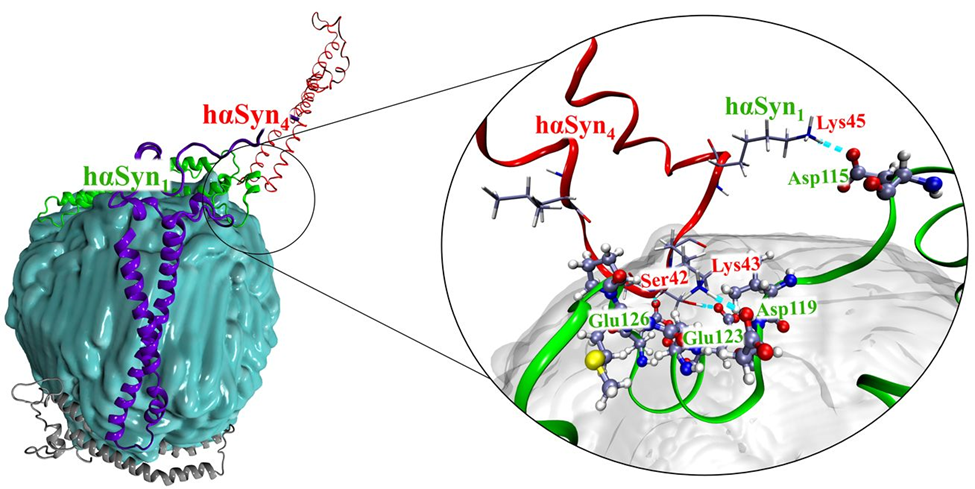
The human α synuclein on polyethylene nanoplastic surface interacting with another human α synuclein molecule
Exploring Macromolecules and Their Modulators with Computational Biology
My educational training in pharmaceutical sciences has helped me to obtain a good understanding of the pharmacokinetics and pharmacodynamics aspects of therapeutic agents. A significant portion of my research focused on systematically studying the structural effects of interactions between protein drug targets and small molecules. Significant outcomes from this line of research are the identification of binding pockets and understanding of the mechanism of drug action by various anticancer drugs, such as merbarone (against human topoisomerase II α), quinacrine as an agonist to TRAIL-DR5 axis, and quinacrine against GLI1-DNA interactions. These pioneering studies have been supported from the experimental evaluations and form the basis of advancements in the drug discovery pipeline.
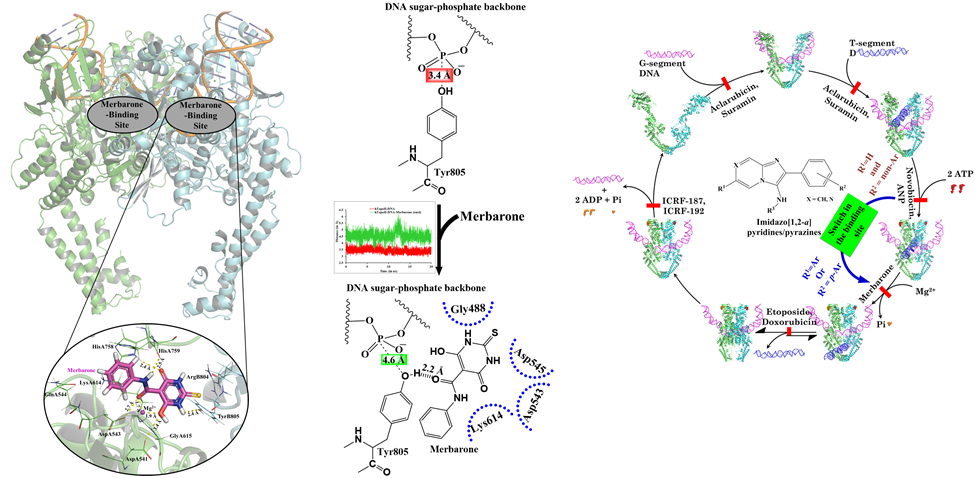
A computational mechanistic evaluation of drug action for the anticancer agent Merbarone (https://doi.org/10.1016/j.jmgm.2018.09.013) and a “choice based switch in the mode of action” upon structural modulation in enzyme’s catalytic cycle (https://doi.org/10.1021/acsmedchemlett.5b00040).
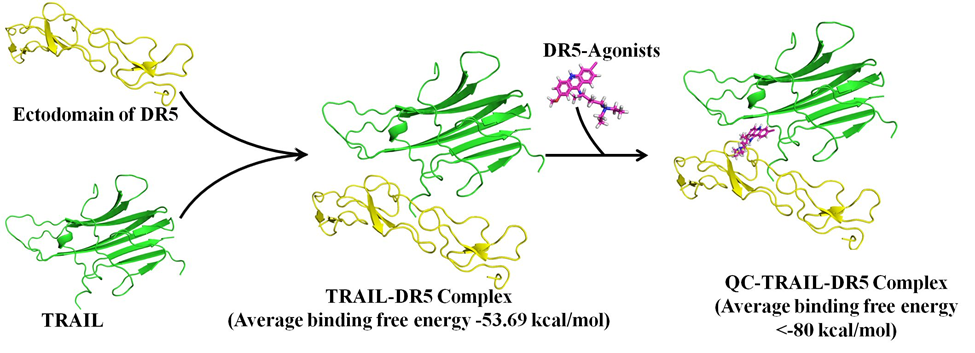
A computational mechanistic evaluation of drug action for the TRAIL-DR5 agonists such as quinacrine (https://doi.org/10.18632/oncotarget.11335), etoposide and doxorubicin (https://doi.org/10.1007/s10495-017-1400-4)
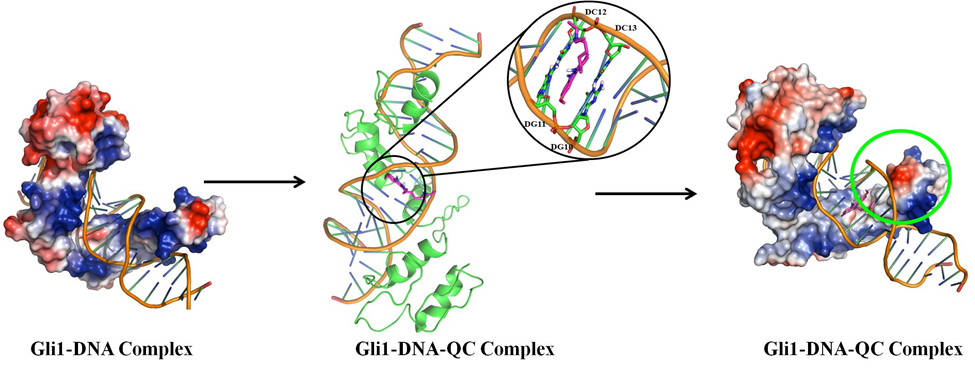
A computational mechanistic evaluation of drug action for the quinacrine on Gli1-DNA axis (https://doi.org/10.1038/srep20600)
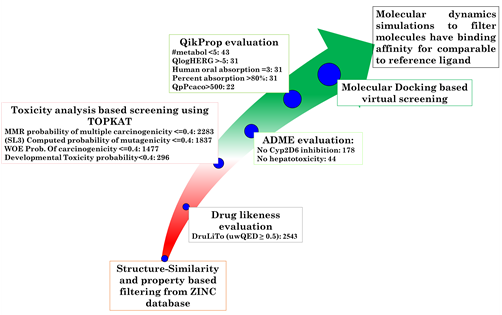
Binding Partner Discovery Using Virtual Screening and Machine Learning
Advancements in computational technologies have revolutionized drug discovery due to numerous advantageous aspects, enabling more efficient identification of potential therapeutic candidates. Virtual screening, combined with machine learning (ML) approaches, provides a powerful platform to explore vast chemical spaces, predict biological activity, and optimize drug-like properties. These techniques allow for rapid prioritization of promising compounds while reducing the cost and time associated with traditional experimental screening. By integrating structural biology, cheminformatics, and ML-driven predictive models, the aim is to accelerate the identification and development of novel therapeutics. This approach ensures a more targeted exploration of candidates with high potential for clinical success, addressing critical health challenges in a data-driven and efficient manner. In this direction, several drug targets were selected and exploited for this purpose during my research career. Several tools and techniques of pharmacoinformatics were employed which resulted into several interesting outcomes. The ML-based efforts for retrobiosynthesis are also undertaken recently.